|
Strain Name
|
C57BL/6N-Tg(CH17-401N15)1Bcgen/Bcgen
|
Common Name
|
B-Tg(hLILRB1/hLILRB4) mice
|
|
Background
|
C57BL/6N
|
Catalog number
|
110878
|
Related Genes
|
LILRB1: ILT2, LIR1, MIR7, PIRB, CD85J, ILT-2, LIR-1, MIR-7, PIR-B
LILRB4: ILT3, LIR5, CD85K, ILT-3, LIR-5
|
NCBI Gene ID
|
11006
|
Protein expression analysis
(LILRB1)
Protein expression analysis (F1)
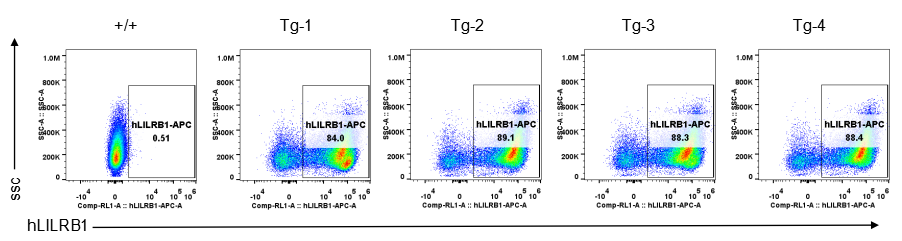
Strain specific LILRB1 expression analysis in transgenic B-Tg(hLILRB1/hLILRB4) mice by flow cytometry. Bone marrow derived dendritic cells from wild-type (+/+) and transgenic B-Tg(hLILRB1/hLILRB4) mice (Tg) were analyzed by flow cytometry with species-specific anti-human LILRB1 antibody. Human LILRB1 was detectable in dendritic cells from B-Tg(hLILRB1/hLILRB4) mice but not wild-type mice.
Protein expression analysis (F2)
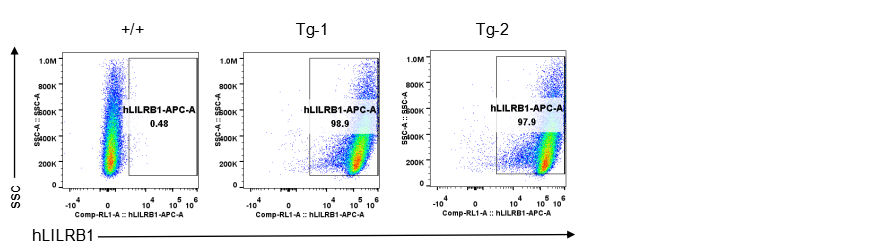
Strain specific LILRB1 expression analysis in transgenic B-Tg(hLILRB1/hLILRB4) mice by flow cytometry. Bone marrow derived dendritic cells from wild-type (+/+) and transgenic B-Tg(hLILRB1/hLILRB4) mice (Tg) were analyzed by flow cytometry with species-specific anti-human LILRB1 antibody. Human LILRB1 was detectable in dendritic cells from B-Tg(hLILRB1/hLILRB4) mice but not wild-type mice.
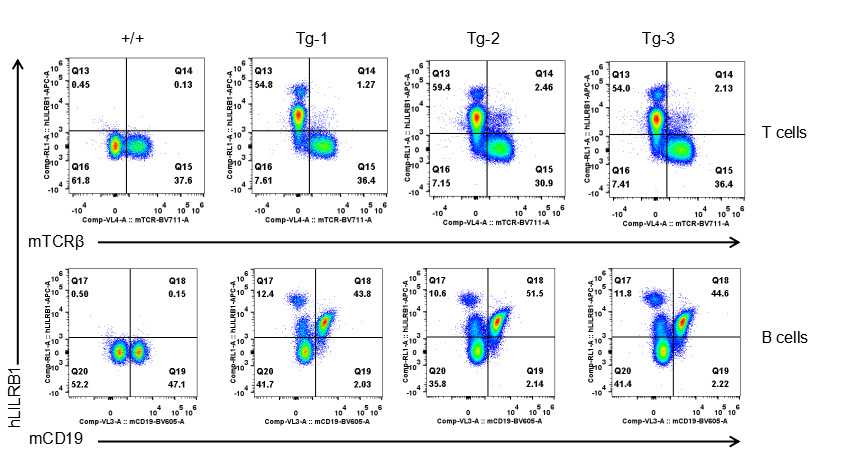
Strain specific LILRB1 expression analysis in transgenic B-Tg(hLILRB1/hLILRB4) mice by flow cytometry. T cells and B cells of blood were collected from wild-type (+/+) and transgenic B-Tg(hLILRB1/hLILRB4) mice (Tg), and analyzed by flow cytometry with species-specific anti-human LILRB4 antibody. Human LILRB1 was both detectable in T cells and B cells from B-Tg(hLILRB1/hLILRB4) mice but not wild-type mice.
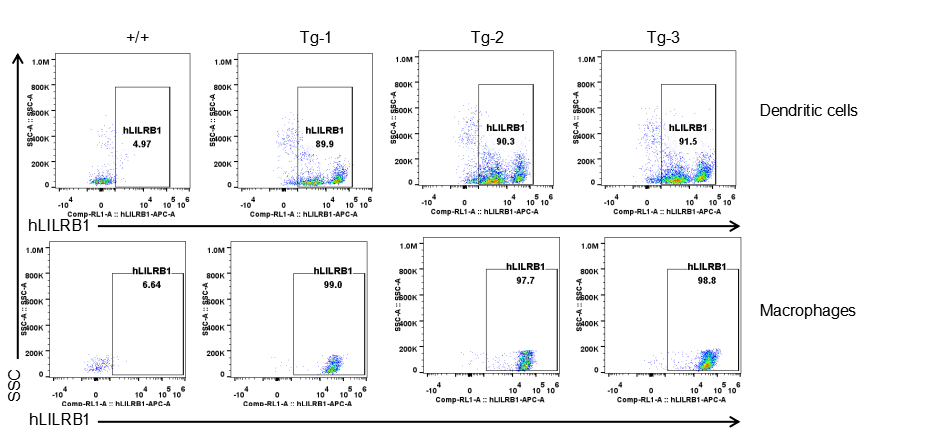
Strain specific LILRB1 expression analysis in transgenic B-Tg(hLILRB1/hLILRB4) mice by flow cytometry. Dendritic cells and macrophages of blood were collected from wild-type (+/+) and transgenic B-Tg(hLILRB1/hLILRB4) mice (Tg), and analyzed by flow cytometry with species-specific anti-human LILRB1 antibody. Human LILRB1 was both detectable in dendritic cells and macrophages from B-Tg(hLILRB1/hLILRB4) mice but not wild-type mice.
Protein expression analysis (F3)
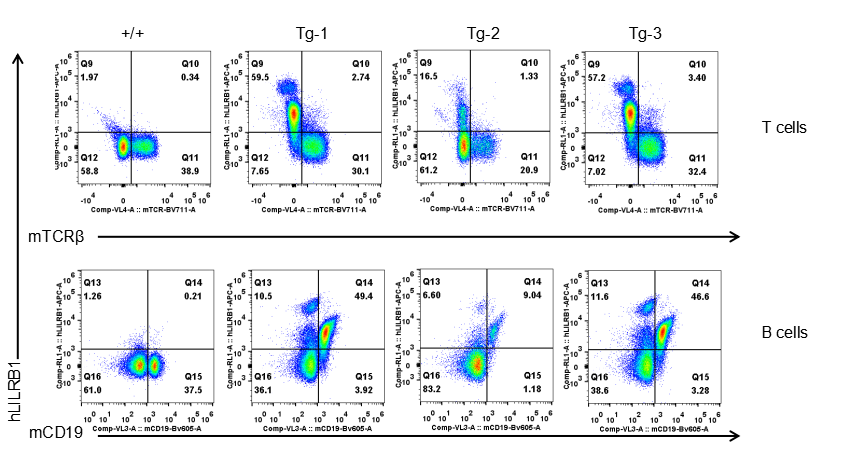
Strain specific LILRB1 expression analysis in transgenic B-Tg(hLILRB1/hLILRB4) mice by flow cytometry. T cells and B cells of blood were collected from wild-type (+/+) and transgenic B-Tg(hLILRB1/hLILRB4) mice (Tg), and analyzed by flow cytometry with species-specific anti-human LILRB4 antibody. Human LILRB1 was both detectable in T cells and B cells from B-Tg(hLILRB1/hLILRB4) mice but not wild-type mice.

Strain specific LILRB1 expression analysis in transgenic B-Tg(hLILRB1/hLILRB4) mice by flow cytometry. Dendritic cells and macrophages of blood were collected from wild-type (+/+) and transgenic B-Tg(hLILRB1/hLILRB4) mice (Tg), and analyzed by flow cytometry with species-specific anti-human LILRB1 antibody. Human LILRB1 was both detectable in dendritic cells and macrophages from B-Tg(hLILRB1/hLILRB4) mice but not wild-type mice.
Protein expression analysis
(LILRB4)
Protein expression analysis (F1)
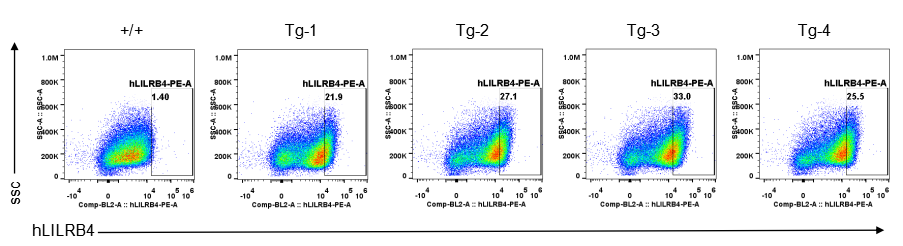
Strain specific LILRB4 expression analysis in transgenic B-Tg(hLILRB1/hLILRB4) mice by flow cytometry. Bone marrow derived dendritic cells from wild-type (+/+) and transgenic B-Tg(hLILRB1/hLILRB4) mice (Tg) were analyzed by flow cytometry with species-specific anti-human LILRB4 antibody. Human LILRB4 was detectable in dendritic cells from B-Tg(hLILRB1/hLILRB4) mice but not wild-type mice.
Protein expression analysis (F2)
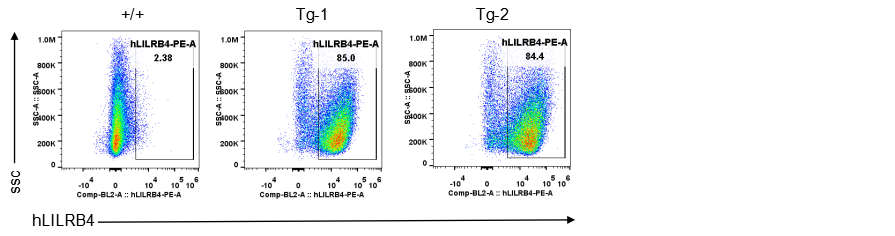
Strain specific LILRB4 expression analysis in transgenic B-Tg(hLILRB1/hLILRB4) mice by flow cytometry. Bone marrow derived dendritic cells from wild-type (+/+) and transgenic B-Tg(hLILRB1/hLILRB4) mice (Tg) were analyzed by flow cytometry with species-specific anti-human LILRB4 antibody. Human LILRB4 was detectable in dendritic cells from B-Tg(hLILRB1/hLILRB4) mice but not wild-type mice.
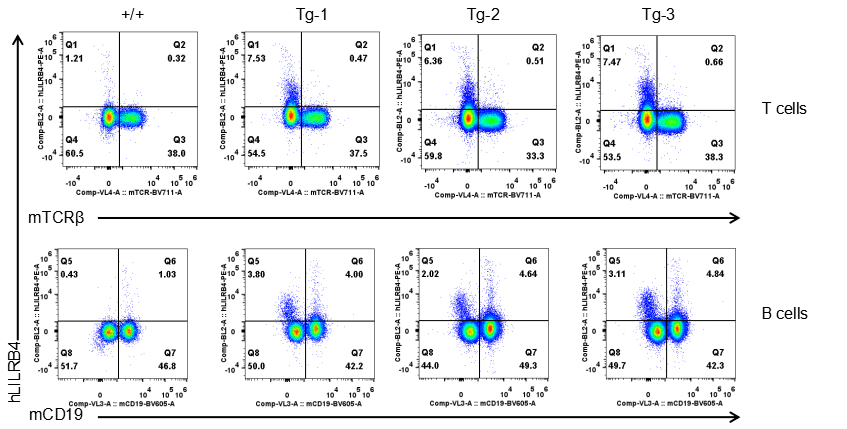
Strain specific LILRB4 expression analysis in transgenic B-Tg(hLILRB1/hLILRB4) mice by flow cytometry. T cells and B cells of blood were collected from wild-type (+/+) and transgenic B-Tg(hLILRB1/hLILRB4) mice (Tg), and analyzed by flow cytometry with species-specific anti-human LILRB4 antibody. Human LILRB4 was not detectable in T cells of B-Tg(hLILRB1/hLILRB4) mice but detectable in B cells.
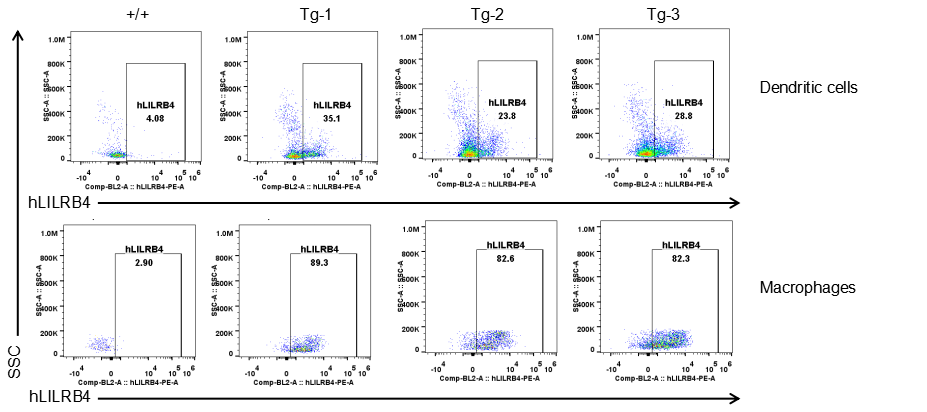
Strain specific LILRB4 expression analysis in transgenic B-Tg(hLILRB1/hLILRB4) mice by flow cytometry. Dendritic cells and macrophages of blood were collected from wild-type (+/+) and transgenic B-Tg(hLILRB1/hLILRB4) mice (Tg), and analyzed by flow cytometry with species-specific anti-human LILRB4 antibody. Human LILRB4 was both detectable in dendritic cells and macrophages from B-Tg(hLILRB1/hLILRB4) mice but not wild-type mice.
Protein expression analysis (F3)
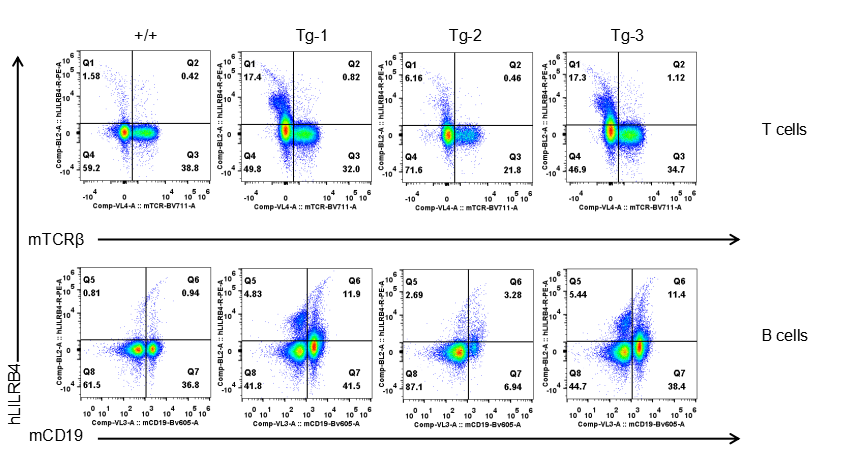
Strain specific LILRB4 expression analysis in transgenic B-Tg(hLILRB1/hLILRB4) mice by flow cytometry. T cells and B cells of blood were collected from wild-type (+/+) and transgenic B-Tg(hLILRB1/hLILRB4) mice (Tg), and analyzed by flow cytometry with species-specific anti-human LILRB4 antibody. Human LILRB4 was not detectable in T cells of B-Tg(hLILRB1/hLILRB4) mice but detectable in B cells.
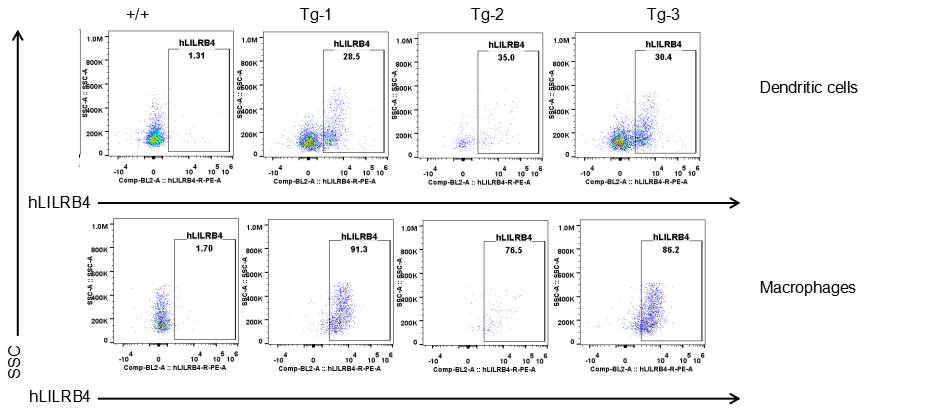
Strain specific LILRB4 expression analysis in transgenic B-Tg(hLILRB1/hLILRB4) mice by flow cytometry. Dendritic cells and macrophages of blood were collected from wild-type (+/+) and transgenic B-Tg(hLILRB1/hLILRB4) mice (Tg), and analyzed by flow cytometry with species-specific anti-human LILRB4 antibody. Human LILRB4 was both detectable in dendritic cells and macrophages from B-Tg(hLILRB1/hLILRB4) mice but not wild-type mice.
Leukocytes cell subpopulation and LILRB4 expression analysis in spleen, thymus, bone marrow and blood of B-Tg(hLILRB1/hLILRB4) mice (non-tumor bearing)
Summary of LILRB4 expression analysis in spleen, thymus, bone marrow and blood of B-Tg(hLILRB1/hLILRB4) mice

Analysis of leukocytes cell subpopulation in spleen
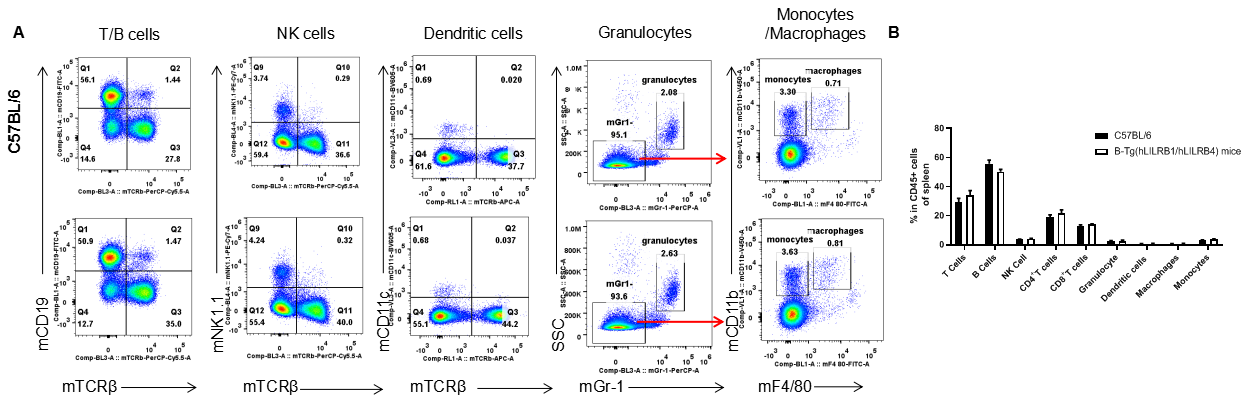
Analysis of spleen leukocyte subpopulations by FACS. Splenocytes were isolated from female C57BL/6 and B-Tg(hLILRB1/hLILRB4) mice (n=3, 8-week-old). Flow cytometry analysis of the splenocytes was performed to assess leukocyte subpopulations. A. Representative FACS plots. Single live cells were gated for the CD45+ population and used for further analysis as indicated here. B. Results of FACS analysis. Percent of T cells, B cells, NK cells, dendritic cells, granulocytes, monocytes and macrophages in B-Tg(hLILRB1/hLILRB4) mice were similar to those in the C57BL/6 mice, demonstrating that LILRB1 and LILRB4 humanized does not change the overall development, differentiation or distribution of these cell types in spleen. Values are expressed as mean ± SEM.
Analysis of T cell subpopulation in spleen
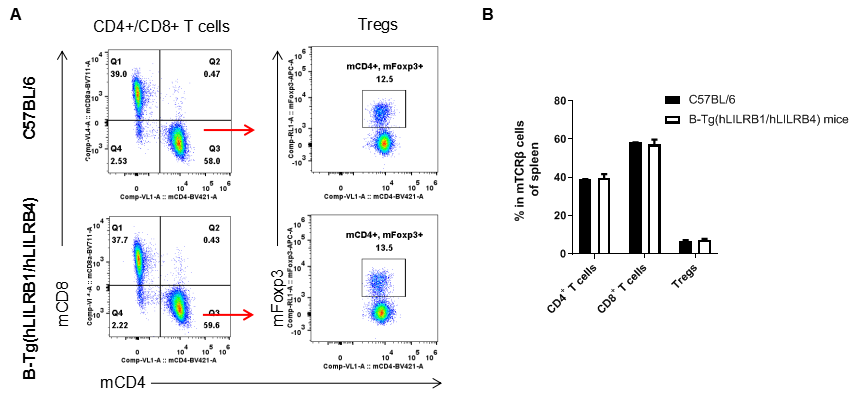
Analysis of spleen T cell subpopulations by FACS. Splenocytes were isolated from female C57BL/6 and B-Tg(hLILRB1/hLILRB4) mice (n=3, 8-week-old). Flow cytometry analysis of the splenocytes was performed to assess leukocyte subpopulations. A. Representative FACS plots. Single live CD45+ cells were gated for TCRβ+ T cell population and used for further analysis as indicated here. B. Results of FACS analysis. The percent of CD4+ T cells, CD8+ T cells and Tregs in B-Tg(hLILRB1/hLILRB4) mice were similar to those in the C57BL/6 mice, demonstrating that introduction of hLILRB1 and hLILRB4 does not change the overall development, differentiation or distribution of these T cell subtypes in spleen. Values are expressed as mean ± SEM.
Analysis of leukocytes cell subpopulation in thymus
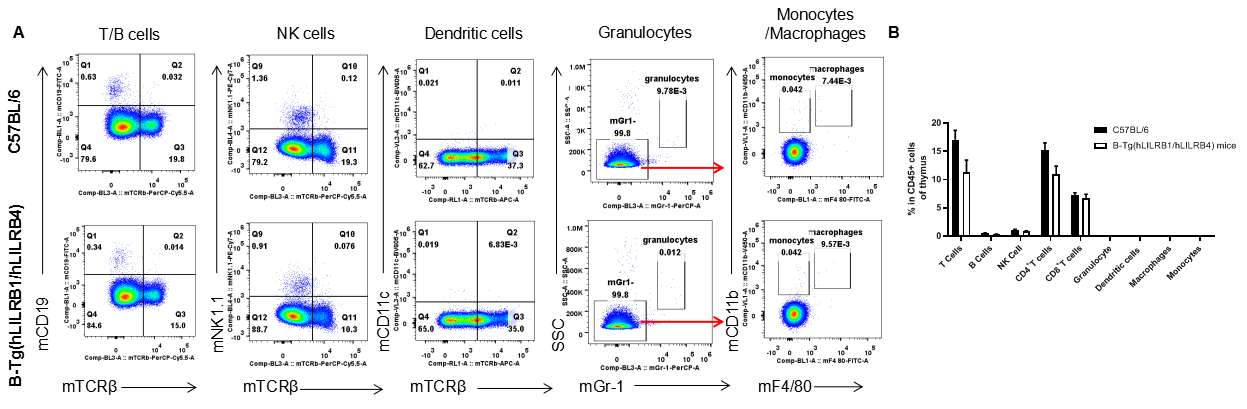
Analysis of thymus leukocyte subpopulations by FACS. Thymocytes were isolated from female C57BL/6 and B-Tg(hLILRB1/hLILRB4) mice (n=3, 8-week-old). Flow cytometry analysis of the thymocytes was performed to assess leukocyte subpopulations. A. Representative FACS plots. Single live cells were gated for the CD45+ population and used for further analysis as indicated here. B. Results of FACS analysis. Percent of T cells, B cells, NK cells, dendritic cells, granulocytes, monocytes and macrophages in B-Tg(hLILRB1/hLILRB4) mice were similar to those in the C57BL/6 mice, demonstrating that LILRB1 and LILRB4 humanized does not change the overall development, differentiation or distribution of these cell types in thymus. Values are expressed as mean ± SEM.
Analysis of T cell subpopulation in thymus
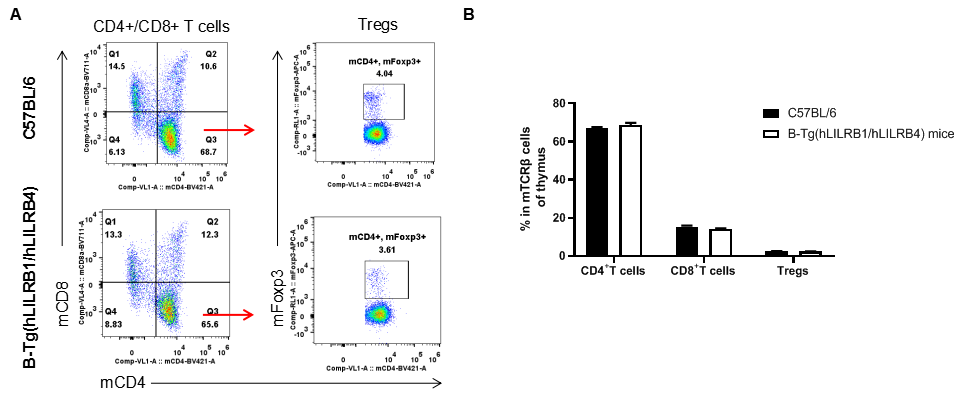
Analysis of thymus T cell subpopulations by FACS. Thymocytes were isolated from female C57BL/6 and B-Tg(hLILRB1/hLILRB4) mice (n=3, 8-week-old). Flow cytometry analysis of the thymocytes was performed to assess leukocyte subpopulations. A. Representative FACS plots. Single live CD45+ cells were gated for TCRβ+ T cell population and used for further analysis as indicated here. B. Results of FACS analysis. The percent of CD4+ T cells, CD8+ T cells and Tregs in B-Tg(hLILRB1/hLILRB4) mice were similar to those in the C57BL/6 mice, demonstrating that introduction of hLILRB1 and hLILRB4 does not change the overall development, differentiation or distribution of these T cell subtypes in thymus. Values are expressed as mean ± SEM.
Analysis of leukocytes cell subpopulation in bone marrow

Analysis of bone marrow leukocyte subpopulations by FACS. Bone marrow leukocyte were isolated from female C57BL/6 and B-Tg(hLILRB1/hLILRB4) mice (n=3, 8-week-old). Flow cytometry analysis of the bone marrow leukocyte was performed to assess leukocyte subpopulations. A. Representative FACS plots. Single live cells were gated for the CD45+ population and used for further analysis as indicated here. B. Results of FACS analysis. Percent of T cells, B cells, NK cells, dendritic cells, granulocytes, monocytes and macrophages in B-Tg(hLILRB1/hLILRB4) mice were similar to those in the C57BL/6 mice, demonstrating that LILRB1 and LILRB4 humanized does not change the overall development, differentiation or distribution of these cell types in bone marrow. Values are expressed as mean ± SEM.
Analysis of T cell subpopulation in bone marrow
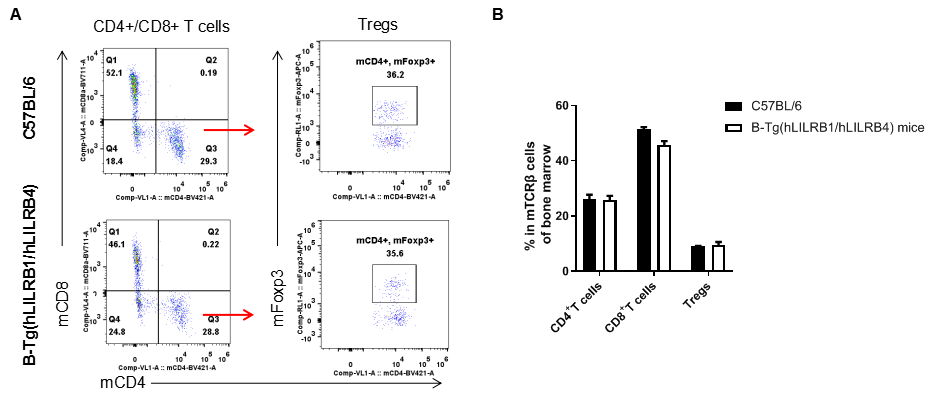
Analysis of bone marrow T cell subpopulations by FACS. Bone marrow leukocyte were isolated from female C57BL/6 and B-Tg(hLILRB1/hLILRB4) mice (n=3, 8-week-old). Flow cytometry analysis of the bone marrow leukocyte was performed to assess leukocyte subpopulations. A. Representative FACS plots. Single live CD45+ cells were gated for TCRβ+ T cell population and used for further analysis as indicated here. B. Results of FACS analysis. The percent of CD4+ T cells, CD8+ T cells and Tregs in B-Tg(hLILRB1/hLILRB4) mice were similar to those in the C57BL/6 mice, demonstrating that introduction of hLILRB1 and hLILRB4 does not change the overall development, differentiation or distribution of these T cell subtypes in bone marrow. Values are expressed as mean ± SEM.
Analysis of leukocytes cell subpopulation in blood

Analysis of blood leukocyte subpopulations by FACS. Blood leukocytes were isolated from female C57BL/6 and B-Tg(hLILRB1/hLILRB4) mice (n=3, 8-week-old). Flow cytometry analysis of the blood leukocytes were performed to assess leukocyte subpopulations. A. Representative FACS plots. Single live cells were gated for the CD45+ population and used for further analysis as indicated here. B. Results of FACS analysis. Percent of T cells, B cells, NK cells, dendritic cells, granulocytes, monocytes and macrophages in B-Tg(hLILRB1/hLILRB4) mice were similar to those in the C57BL/6 mice, demonstrating that LILRB1 and LILRB4 humanized does not change the overall development, differentiation or distribution of these cell types in blood. Values are expressed as mean ± SEM.
Analysis of T cell subpopulation in blood
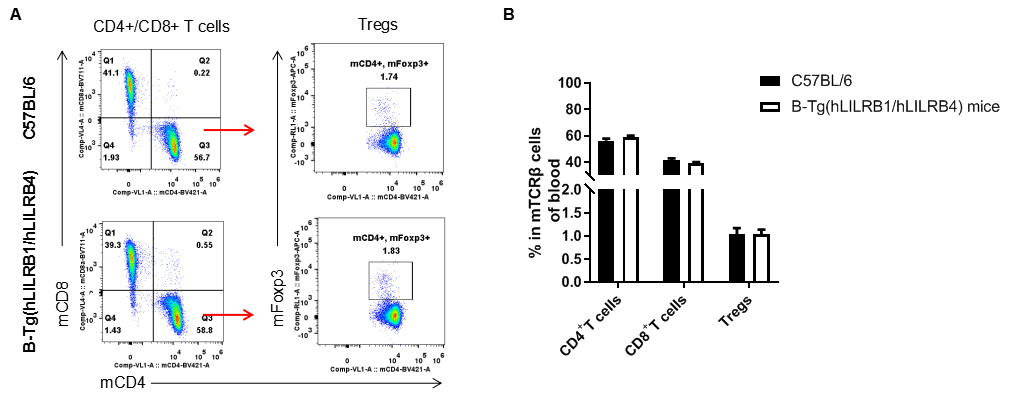
Analysis of blood T cell subpopulations by FACS. Blood leukocytes were isolated from female C57BL/6 and B-Tg(hLILRB1/hLILRB4) mice (n=3, 8-week-old). Flow cytometry analysis of the blood leukocytes were performed to assess leukocyte subpopulations. A. Representative FACS plots. Single live CD45+ cells were gated for TCRβ+ T cell population and used for further analysis as indicated here. B. Results of FACS analysis. The percent of CD4+ T cells, CD8+ T cells and Tregs in B-Tg(hLILRB1/hLILRB4) mice were similar to those in the C57BL/6 mice, demonstrating that introduction of hLILRB1 and hLILRB4 does not change the overall development, differentiation or distribution of these T cell subtypes in blood. Values are expressed as mean ± SEM.
Blood chemistry of B-Tg(hLILRB1/hLILRB4) mice
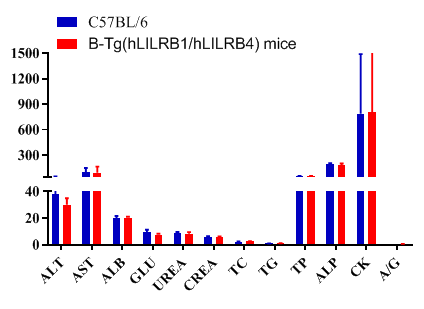
Blood chemistry tests of B-Tg(hLILRB1/hLILRB4) mice. Serum from the C57BL/6 and B-Tg(hLILRB1/hLILRB4) mice (n=8, 9 week-old) was collected and analyzed for levels of indicators. The measurements of B-Tg(hLILRB1/hLILRB4) mice were similar to that in C57BL/6 mice, indicating that humanization does not change the health of related tissues, such as liver. Values are expressed as mean ± SEM.
Blood routine test in B-Tg(hLILRB1/hLILRB4) mice
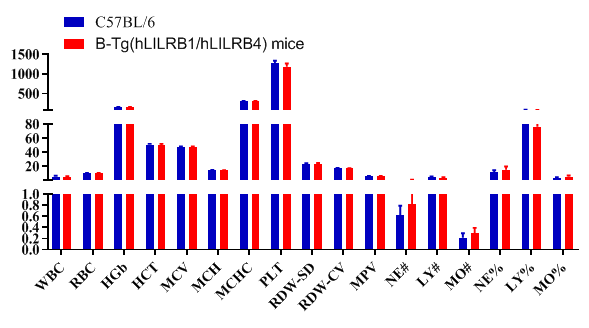
Complete blood count (CBC). Blood from female C57BL/6 and B-Tg(hLILRB1/hLILRB4) mice (n=8, 9 week-old) was collected and analyzed for CBC. The measurements of B-Tg(hLILRB1/hLILRB4) mice were similar to that in C57BL/6 mice, indicating that humanization does not change blood cell composition and morphology. Values are expressed as mean ± SEM.
Relative LILRB4 mRNA expression in B-Tg(hLILRB1/hLILRB4) mice
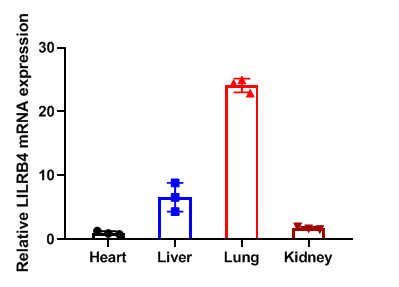
Relative LILRB4 mRNA expression. Heart、liver、lung and kidney from female C57BL/6 and B-Tg(hLILRB1/hLILRB4) mice (n=3, 9 week-old) was collected and analyzed for q-PCR. Human LILRB4 mRNA was detectable in heart、liver、lung and kidney from B-Tg(hLILRB1/hLILRB4) mice but not wild-type mice. Values are expressed as mean ± SEM.
Tumor growth curve & Body weight changes
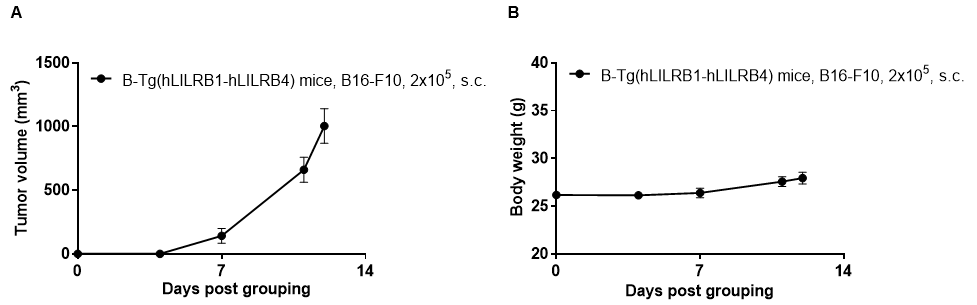
Subcutaneous homograft tumor growth of B16-F10 cells. B16-F10 cells (2x105) were subcutaneously implanted into B-Tg(hLILRB1/hLILRB4) mice (female, 10-11 week-old, n=5). Tumor volume and body weight were measured twice a week. (A) Average tumor volume ± SEM. (B) Body weight (Mean± SEM). Volume was expressed in mm3 using the formula: V=0.5 X long diameter X short diameter2. As shown in panel A, B-Tg(hLILRB1/hLILRB4) mice were able to establish tumors in vivo and can be used for efficacy studies.
Analysis of tumor infiltrating lymphocytes (TILs)
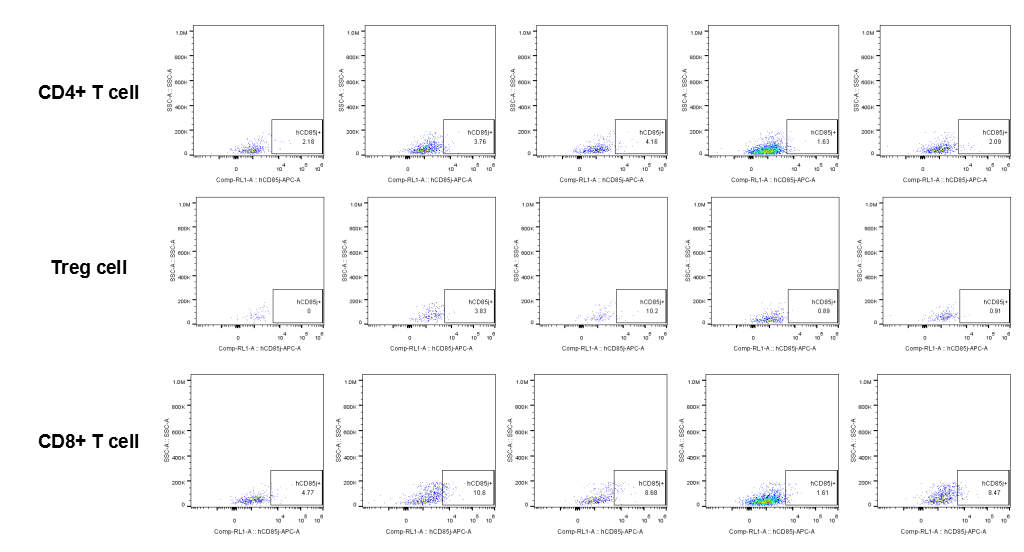
The expression of hLILRB1 in CD4+ T cell, Treg cell and CD8+ T cell.Tumor cells were harvested at the endpoint of experiment, and the lymphocytes were analyzed by flow cytometry.
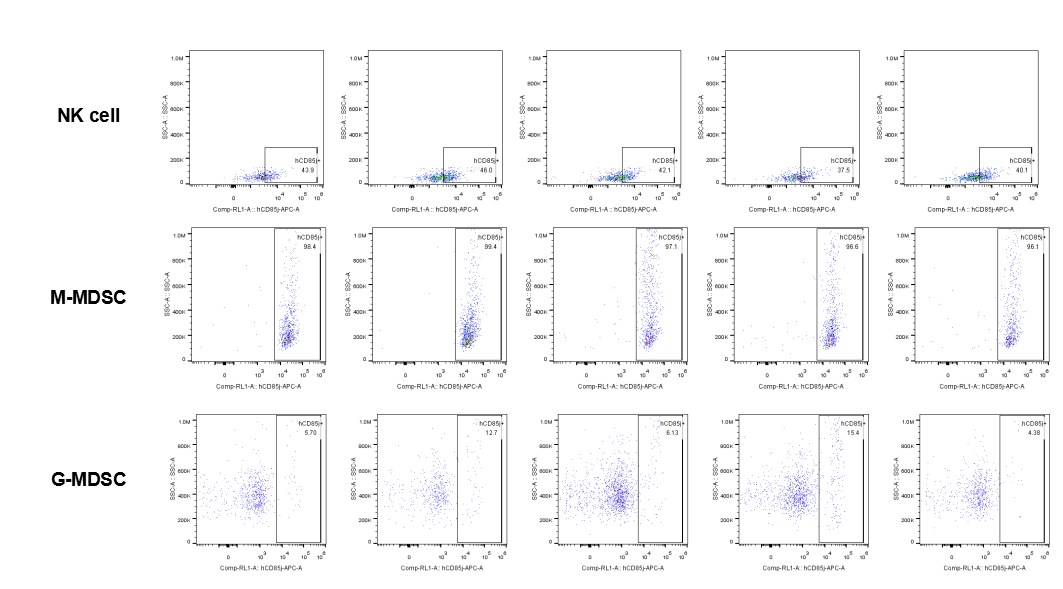
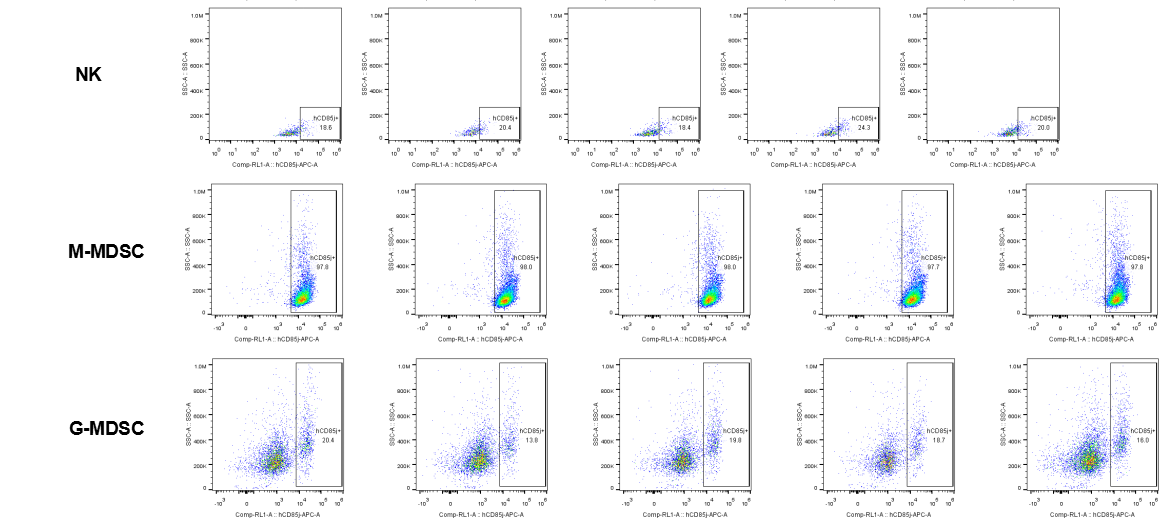
The expression of hLILRB1 in NK and MDSC (G-MDSC, M-MDSC).Tumor cells were harvested at the endpoint of experiment, and the lymphocytes were analyzed by flow cytometry.
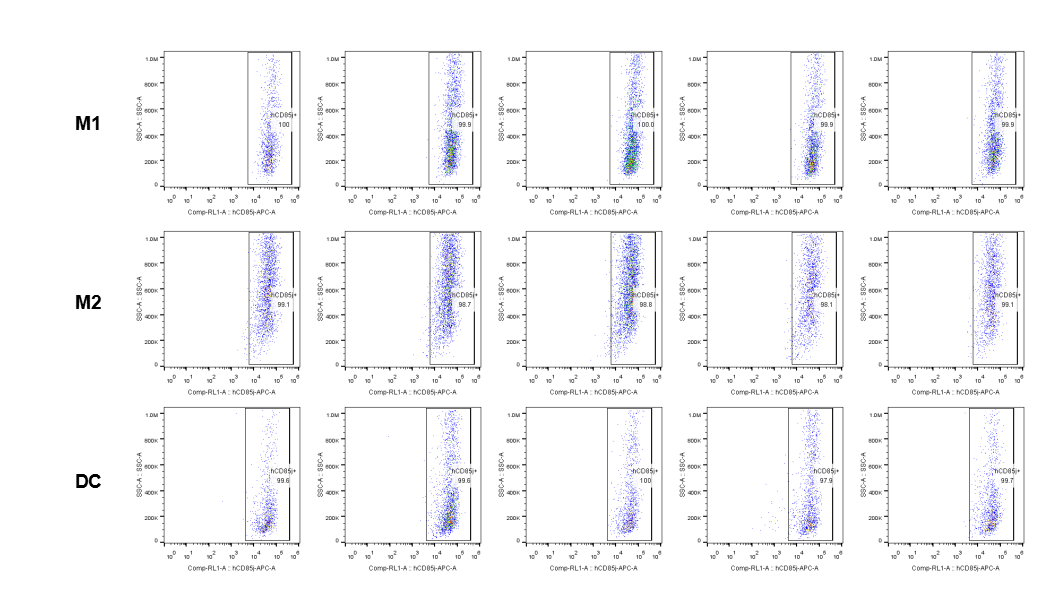
The expression of hLILRB1 in macrophage (M1, M2) and DC.Tumor cells were harvested at the endpoint of experiment, and the lymphocytes were analyzed by flow cytometry.
The expression of hLILRB4 in CD4+ T cell, Treg cell and CD8+ T cell.Tumor cells were harvested at the endpoint of experiment, and the lymphocytes were analyzed by flow cytometry.
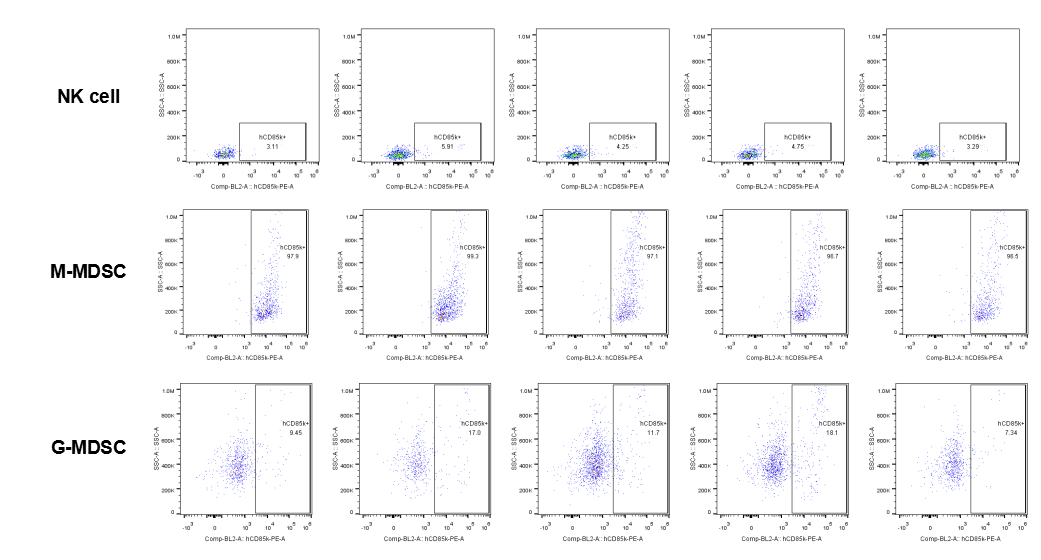
The expression of hLILRB4 in NK cell and MDSC (G-MDSC, M-MDSC).Tumor cells were harvested at the endpoint of experiment, and the lymphocytes were analyzed by flow cytometry.
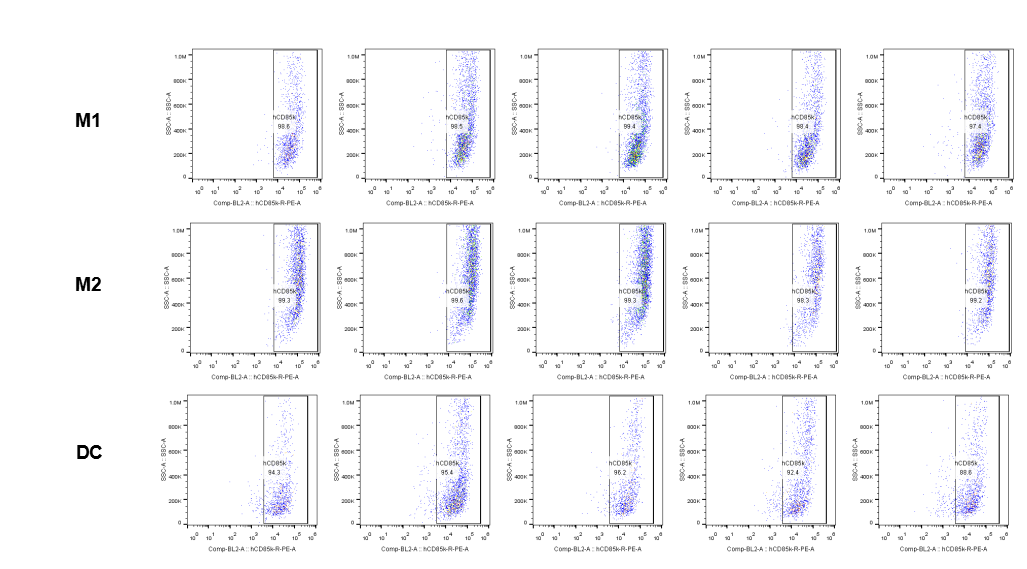
The expression of hLILRB4 in macrophage (M1, M2) and DC.Tumor cells were harvested at the endpoint of experiment, and the lymphocytes were analyzed by flow cytometry.
Tumor growth curve & Body weight changes
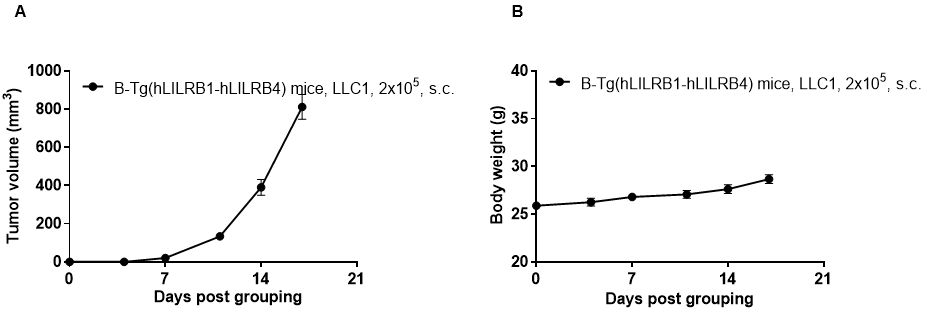
Subcutaneous homograft tumor growth of LLC1 cells. LLC1 cells (2x105) were subcutaneously implanted into B-Tg(hLILRB1/hLILRB4) mice (female, 10-11 week-old, n=5). Tumor volume and body weight were measured twice a week. (A) Average tumor volume ± SEM. (B) Body weight (Mean± SEM). Volume was expressed in mm3 using the formula: V=0.5 X long diameter X short diameter2. As shown in panel A, B-Tg(hLILRB1/hLILRB4) mice were able to establish tumors in vivo and can be used for efficacy studies.
Analysis of tumor infiltrating lymphocytes (TILs)
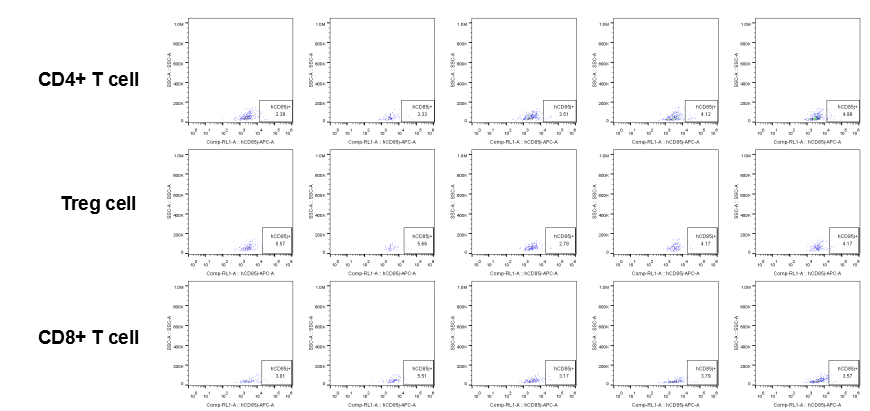
The expression of hLILRB1 in CD4+ T cell, Treg cell and CD8+ T cell.Tumor cells were harvested at the endpoint of experiment, and the lymphocytes were analyzed by flow cytometry.
The expression of hLILRB1 in NK cell and MDSC (G-MDSC, M-MDSC).Tumor cells were harvested at the endpoint of experiment, and the lymphocytes were analyzed by flow cytometry.
The expression of hLILRB1 in macrophage (M1, M2) and DC.Tumor cells were harvested at the endpoint of experiment, and the lymphocytes were analyzed by flow cytometry.
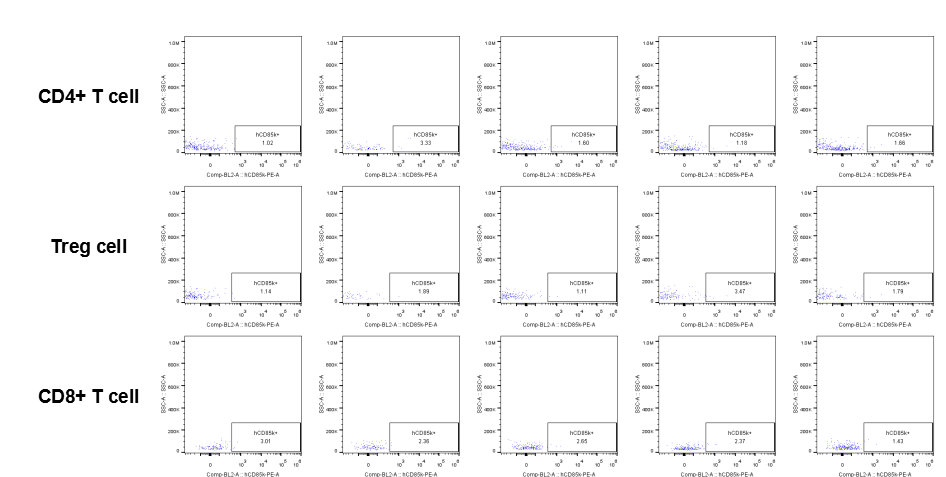
The expression of hLILRB4 in CD4+ T cell, Treg cell and CD8+ T cell.Tumor cells were harvested at the endpoint of experiment, and the lymphocytes were analyzed by flow cytometry.
The expression of hLILRB4 in NK cell and MDSC (G-MDSC, M-MDSC).Tumor cells were harvested at the endpoint of experiment, and the lymphocytes were analyzed by flow cytometry.
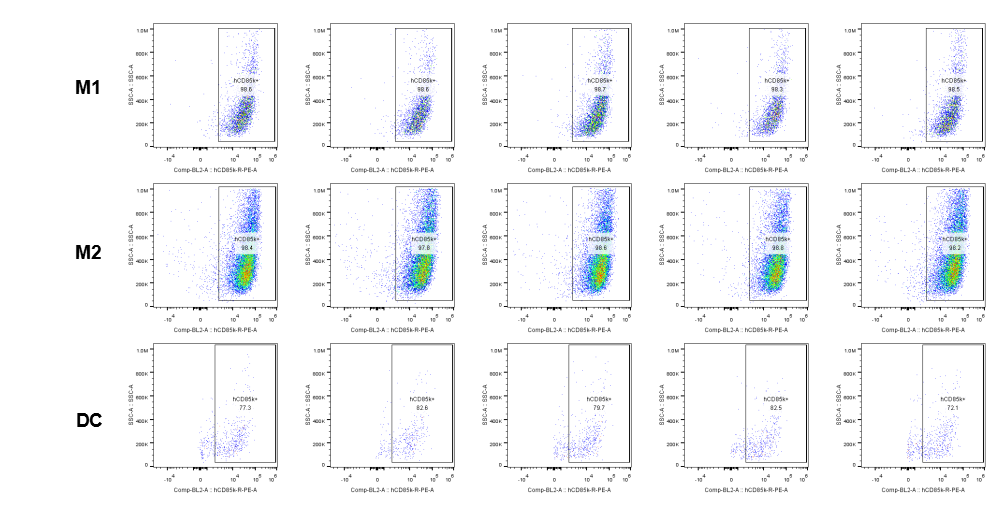
The expression of hLILRB4 in macrophage (M1, M2) and DC.Tumor cells were harvested at the endpoint of experiment, and the lymphocytes were analyzed by flow cytometry.
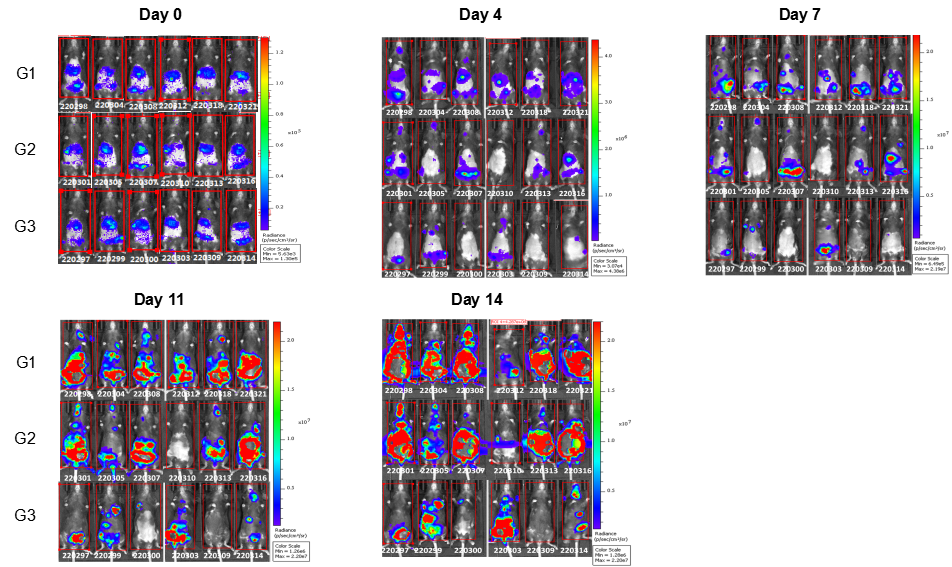
In vivo efficacy of anti-human LILRB4 antibody

Antitumor activity of anti-human LILRB4 antibody in B-Tg(hLILRB1/hLILRB4) mice. (A) Anti-human LILRB4 antibody inhibited B-hLILRB4 luc EL4 lymphoma growth in B-Tg(hLILRB1/hLILRB4) mice. B-hLILRB4 luc EL4 cells (2x105) were injected by tail vein into B-Tg(hLILRB1/hLILRB4) mice (female, 6 week-old, n=6). Mice were grouped when total flux reached approximately 106 Ig, and treated with anti-hLILRB4 antibody in panel A. (B) Body weight changes during treatment. As shown in panel A, anti-hLILRB4 antibody IO-202(in house) was efficacious in controlling tumor growth in B-Tg(hLILRB1/hLILRB4) mice. Values are expressed as mean ± SEM.

















































 京公网安备:
京公网安备: